What's new in CodonCode Aligner 4
CodonCode Aligner version 4.0 introduces support for Next Generation Sequencing and several new features and improvements like a new contig overview, building trees for selections, splitting contigs using trees, highlighting sequences with labels, reading and writing new file formats, importing subsets, and much more.
CodonCode Aligner 4.1 added new filters for the difference table and the option to switch between a stacked and packed overview for aligned samples. It is comatible with Windows 8 and OS X Mountain Lion (10.8).
CodonCode Aligner 4.2 introduces support for 64-bit Windows. On 64-bit versions of Windows, CodonCode Aligner 4.2.1 can now use memory beyond the 1.4 GB limit on 32-bit Windows, up to the available amount of physical memory (RAM).
|
|
Improved Overview
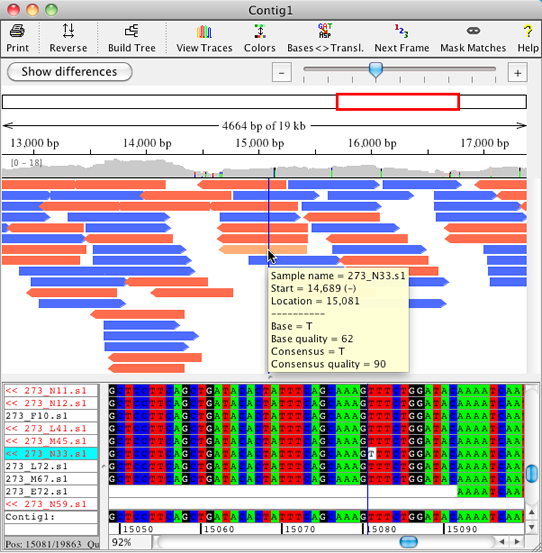
The new overview for contigs in CodonCode Aligner was optimized for large projets. An improved display of the sample arrows allows the user to get a better idea of gaps and sample position in alignments. Mouse overs show significant information for each position, and when zooming in on the samples, discrepancies are shown in colors on the sample arrows.
The overview allows for zooming with a zoom scale and shows which range is displayed.
The coverage panel above the sample arrows shows colored discrepancies (with a settable threshold) and allows for fast navigation to any region of interest. |
Additional Tree Functionalities
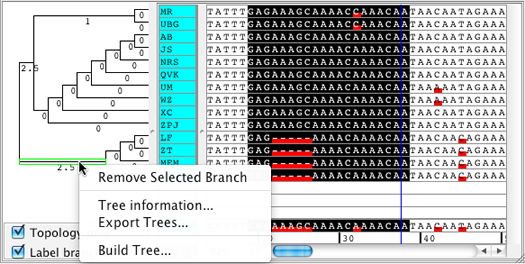
Build Neighbor-Joining trees for selected bases only which enables the user to sort samples by similarity in a selected region of the contig.
Contigs can be split into two new contigs by removing a branch of their tree.
Additional options when building Neighbor-Joining trees to include internal gaps or all gaps when calculating differences. |
 |
Highlighting sequences
Highlight your sequences, contigs and manual folders in the project view with colored labels:

|
Next Generation Sequencing support
CodonCode Aligner now also has support for anlayzing & viewing NGS data. The new overview allows to easily work with larger projects and makes it easier to see how your sequences align and where they differ. The new project format allows for a much larger amount of data and is faster when reading and writing your projects. New file formats like fastq, and sam files enable users to import and export their data in common formats. Importing subsets allows to import only sequences that share a certain amount of words with selected sequences in your project, thus enabling the user to import only certain samples from a large dataset. And assemblies and alignments were adapted to work with more sequences and larger contigs. |
More...
- Import a subset of samples: Import every nth sequence or only those samples that match the selected sequences in your project.
- Read & write fastq files: Import and export your sequences in fastq format.
- Read sam files: Import your sequences in sam format.
- Add tags to all samples: Add the same tag to all samples of a contig at the same time.
- New project format: CodonCode Aligner now saves your projects in one file, thus being faster and more memory efficient when reading and writing many sequences.
- Tip of the Day: Look at the "Tip of the Day" in the Help menu to find out about different functionalities in CodonCode Aligner.
- Support for larger projects
- Assemble and align Next Generation Sequencing data
- Improved speed and memory use during assemblies and alignments
CodonCode Aligner 4.1 is compatible with Windows 8 and OS X Mountain Lion (10.8).
Interested? Watch the movie showing the new features of CodonCode Aligner 4.